Prediction of High Performance in Sports and Games based on Genetic analysis
Download PDF
Champions are born with inherited physical potential, and by providing all kinds of support like technical, psychological, environmental, they can be molded into top class performers. Individuals have always been screened through different means for their athletic, physical and behavioral prowess that contributes to their high performance in sports. There has been a variety of screening methods like Pre-Participation Physical Evaluation (PPE) to screen for illnesses, injury susceptibility or medical conditions. Counseling has always been done to understand the psychological aspect of the athletes. This has been a mandatory administrative requirement for sports authorities/federation for organized sports for years. However, the main drawback to these screening methods have always been that it was time-consuming, non-uniformity of protocols, compliance issues and occasionally no follow-up is done. With all these existing conventional screening procedures, if the athlete does not possesses their required inherited potentialities any form of support will result in identifying/developing an average athlete.
Prediction of High Performance in Sports and Games based on Genetic analysis
1Rushali Kamath, 1,2Bannikuppe S. Vishwanath, and 3*Mallaiah Chandrakumar
1Department of Studies in Biochemistry, 2Department of Studies in Molecular Biology 3Department of Studies in Physical Education and Sports Science, University of Mysore, Manasagangotri, Mysuru – 570 005.
*Correspondence Author
- Introduction
Champions are born with inherited physical potential, and by providing all kinds of support like technical, psychological, environmental, they can be molded into top class performers. Individuals have always been screened through different means for their athletic, physical and behavioral prowess that contributes to their high performance in sports. There has been a variety of screening methods like Pre-Participation Physical Evaluation (PPE) to screen for illnesses, injury susceptibility or medical conditions. Counseling has always been done to understand the psychological aspect of the athletes. This has been a mandatory administrative requirement for sports authorities/federation for organized sports for years. However, the main drawback to these screening methods have always been that it was time-consuming, non-uniformity of protocols, compliance issues and occasionally no follow-up is done. With all these existing conventional screening procedures, if the athlete does not possesses their required inherited potentialities any form of support will result in identifying/developing an average athlete.
Is there any other way a standardized protocol could be established for identification of sports high-performances in athletes that can screen for prevalent conditions, is accurate, practical, and cost-effective and can identify treatable conditions? A bonus would be that it should be non-invasive and simple.
The answer to this lies in a concept of molecular biology where a simple saliva sample or cheek swab can donate the whole genetic blueprint of the person. Here is where basic molecular techniques of extraction of DNA meet genomic studies. Sports Genomics is now a new upcoming field that is gaining momentum in the world of analysis and improvement of sports high performance and uses the genetic knowledge of the person to assess their abilities and performances in sports.
Athlete status is now found to be a heritable trait and on an average 66% of variance in athlete status is explained by genetic factors. Its development is later done by environmental factors (1). There are multitudes of factors that are responsible for elite athlete performance that is a combination of physiological and psychological factors and they can be measured by mapping the genetic markers. The traits that are analyzed are maximum oxygen uptake (VO2max), skeletal muscle composition, and motor activity and sub maximum aerobic performance. Performance variants can be found in differences of lifestyles, environment conditions like terrain, distance from sea level, eating habits, motivation, sports equipment and training programs and is all contained within the genetic material of an individual.
Assessing the genetic trait can now be done by simple biochemical methods and proper gene analysis can be done to generate a blueprint of the athletes’ traits which will be advantageous in a wide variety of ways. A person can be screened for their genetic disabilities beforehand preventing avoidable fatalities and health complications. Knowing whether the athlete bears favorable traits for the field of their interest helps the coach to effectively train them and the best diet, training regime and personalized psychological counseling could be tailor-made for the athlete in question.
This is the next step of enhancing athletic performance and is done in many developed nations already. It is high time this concept was incorporated in the Indian sports fields to generate elite athletes more effectively.
- PRINCIPLE
2.1 Genome and Chromosomes
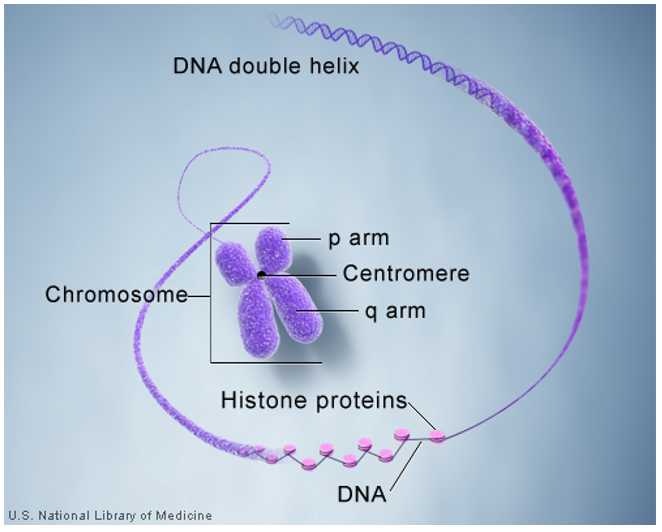
|
Fig 1: Condensation of DNA, subsequent wrapping around histone proteins and formation of chromosomes. (https://ghr.nlm.nih.gov/primer/basics/chromosome) |

The human genome represents the full complement of genetic material in a human cell. The genome is distributed among 23 pairs of chromosomes and it is this that imparts our individual uniqueness. Chromosomes are made up of genes that contain the genetic blueprint or “roadmap” of instructions for almost every aspect of our being.
Genes are traits of heredity that consists of a particular set of instructions that determine the features the organism possesses, how it behaves and how it survives. They are made of DNA that encode for all proteins within the body be it enzymes, structural proteins or even cellular machinery by the process of transcription and translation. (33)
|
Fig 2: 23 pairs of chromosomes in humans. (https://ghr.nlm.nih.gov/primer/basics/howmanychromosomes) |
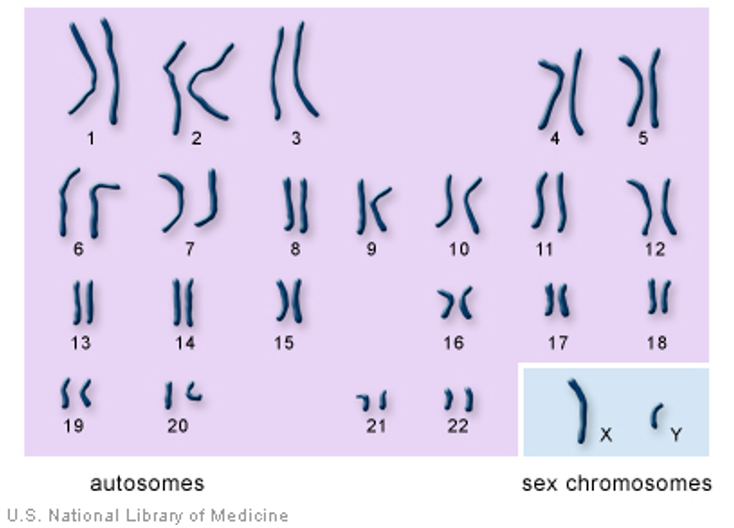
Humans have 23 pairs of chromosomes with each set inherited from one parent. Together the 23 pairs of chromosomes encode all the traits and characters of humans. The Human Genome Project has mapped the locations of all the genes and now it is possible to locate the specific gene of interest.
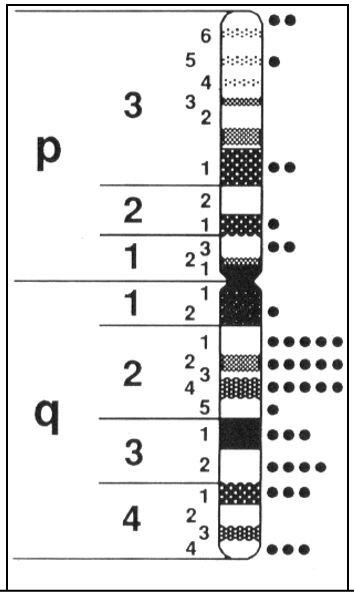
|
Fig 3: Banding pattern of the chromosomes. The arm names can be p or q. within the arms, numbering is often applied to denote the position of genes. (https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/g-banding) |
The chromosomes were visualized at the beginning because they stained vividly. Using a variety of stains, the stained bands were observed and the genes could be located easily. Locations of genes could be traced and it was called locus. The location of a trait (characteristic of a gene) was found to be mostly conserved across all members of a species. Egs: Sickle cell anemia mutation holders always bear the mutation in the 11th chromosome in p arm at 15th band at 5th position. So the chromosome locus assigned to that gene is given as Chr11p15.5. This is how the location of the genes can be mapped and traced.
2.2 Central Dogma of Molecular Biology- DNA to proteins.
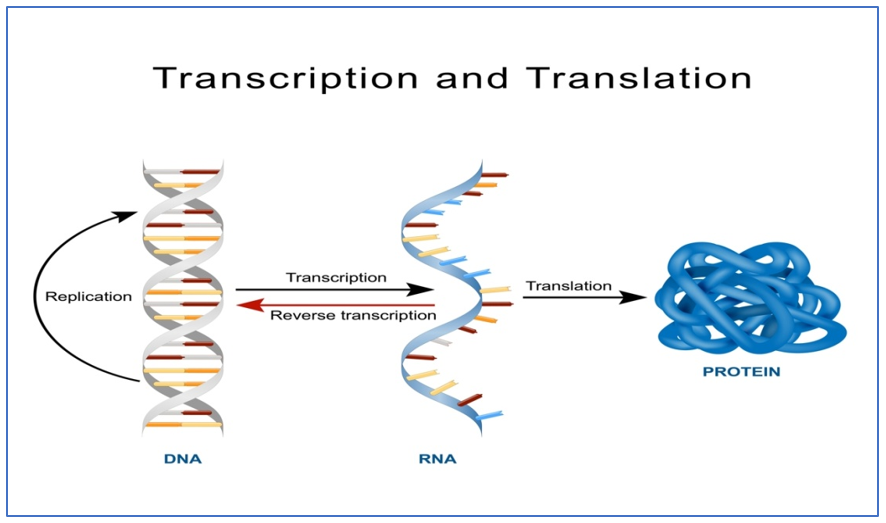
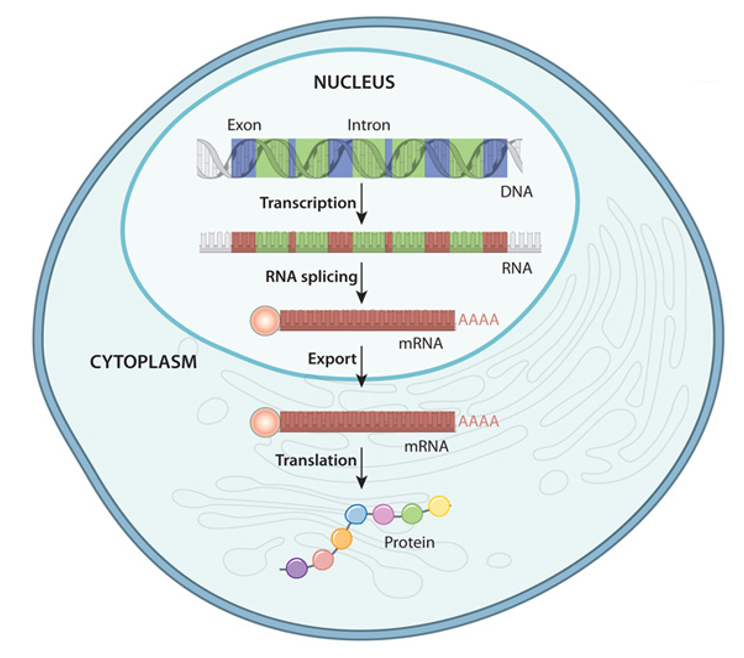
|
Fig 4: Above: https://biologywise.com/protein-synthesis-process. Below: https://www.nature.com/scitable/topicpage/gene-expression-14121669/ |
The DNA that makes up the chromosomes are in the form of a nucleotide sequence. This nucleotide sequence gets transcripted to mRNA where the junk DNA is eliminated and only the genes are gathered as RNA and it moves out of the nucleus, gets translated to form polypeptides that eventually make up the protein of interest.
2.3 Formation of Proteins:
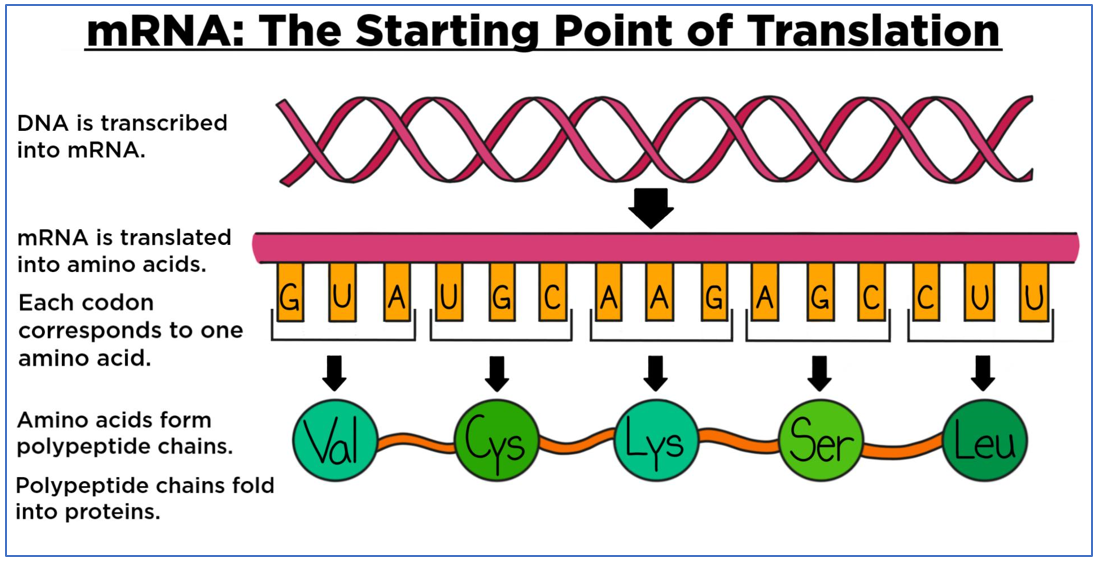
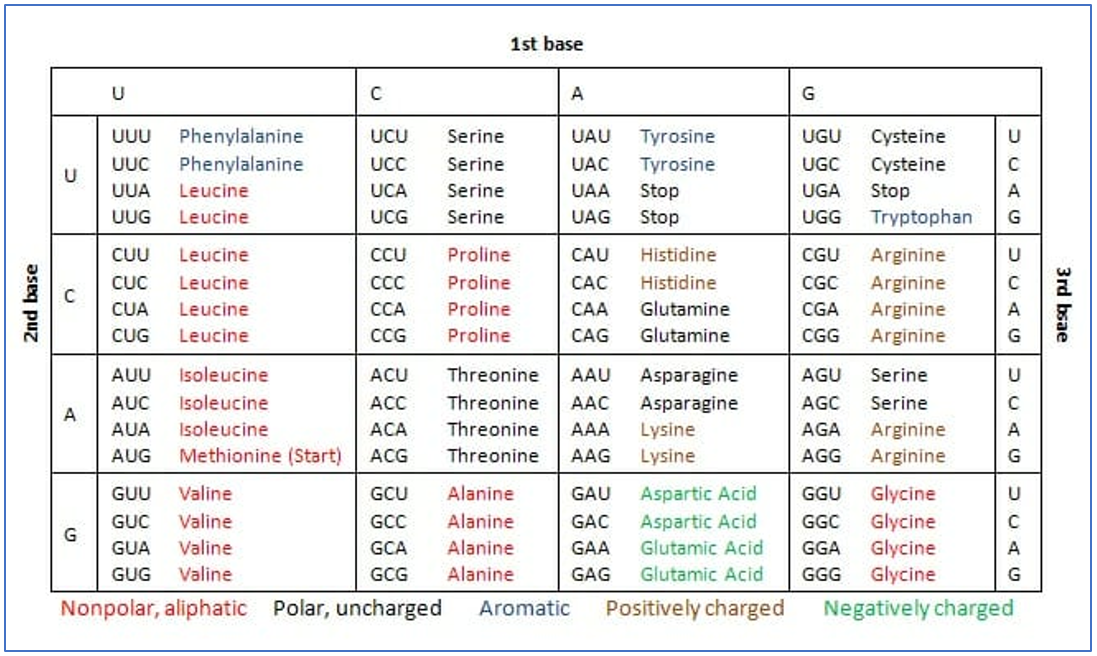
|
Fig 5: Above: The mRNA translation process overview. (https://www.expii.com/t/role-of-mrna-10218). Below: The genetic code with names of amino acids encoded and nature of amino acids. (https://biologydictionary.net/genetic-code/) |
The sequence of nucleotides on the mRNA is translated in the form of codons. Three nucleotides encode one amino acid. The amino acids encoded are listed in the table (Fig:5, below) and can be polar, non-polar, hydrophilic and hydrophobic.
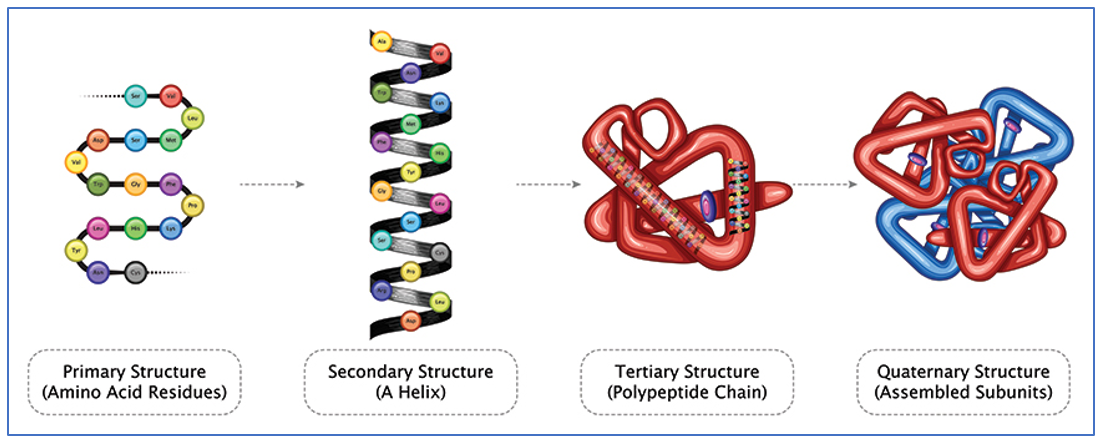
|
Fig 6: https://www.atascientific.com.au/3-protein-analysis-techniques/ |
The polypeptide folds and forms a 3D structure from the linear structure with all the hydrophobic residues internalized and hydrophilic residues outside to form a stable functional structure. The protein formed at stable and in most functional state. All proteins formed in the body folds this way: be it muscle proteins, enzymes, hemoglobin, etc. Any variations in the nucleotides will hence be carried unto the mRNA and will affect the amino acid residues and eventually change the protein structure and function.
This can be exemplified by the disorder of Sickle Cell Anemia.
2.4 Sickle Cell Anemia
|
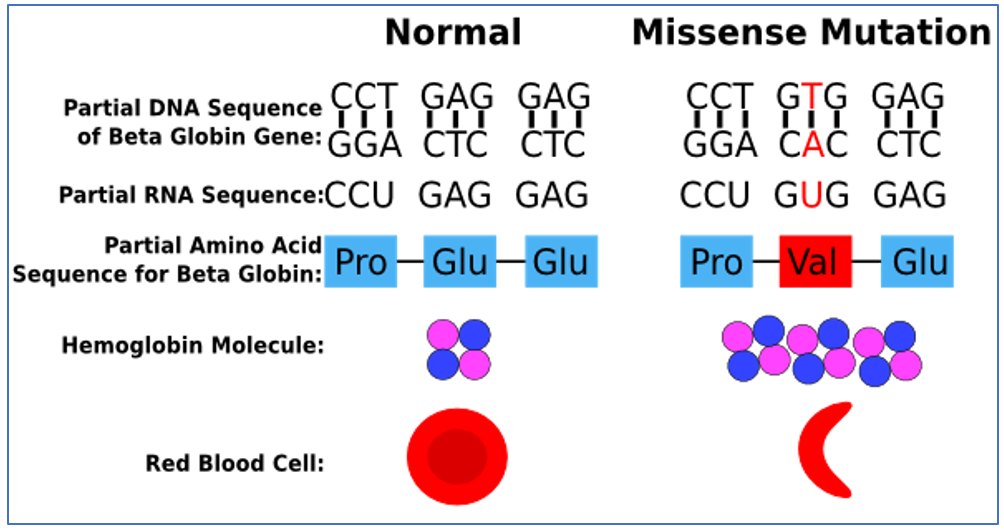
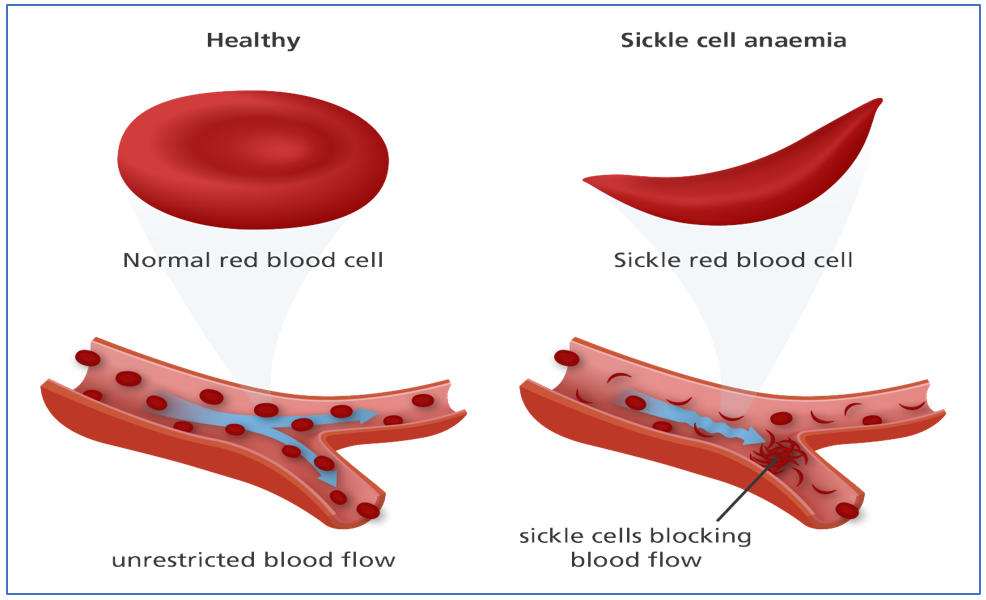
Fig7: Above: A single nucleotide substitution A to T, leads to the formation of another amino acid residue: Glutamate to Valine. Below: This substitution causes a whole other change in the structure thus leading to sickling and polymerization of hemoglobin, causing haemorrhage. |
Sickle Cell Anemia is a genetic disorder of a single nucleotide substitution (A to T) in the HBB gene that encodes hemoglobin. Hemoglobin is a crucial element of RBCs and plays an important role in the distribution of oxygen across the body. Any variations in the hemoglobin can have adverse conditions, especially in hypoxic conditions. This replacement of A by T, leads to the formation of Valine instead of Glutamic Acid. Glutamic acid is present on the outer part of hemoglobin protein but if it gets replaced by the hydrophobic valine, that position of amino acid will distort and move to the core and the whole structure of hemoglobin will change.
This is a genetic mutation; hence the RBCs all across the body will have defective hemoglobin. This defective hemoglobin sickles or polymerizes in hypoxic conditions thus will not be able to provide oxygen effectively to the body parts, which can be fatal. This condition is not detectable in normoxia. In fact, 20-year-old Bennie F. Abram who had started his first day of football practice as a junior at the University of Mississippi collapsed and died. In 2010, 20-year-old Jospin Milandu and 15-year-old Oliver Louis also died unexpectedly while working out with their teams due to this.
Hence, determining this by gene sequence analysis became a necessity and is now in practice. This is a basic overview of how a single nucleotide change can influence body proteins.
2.5 Single Nucleotide Polymorphisms (SNPs)
|
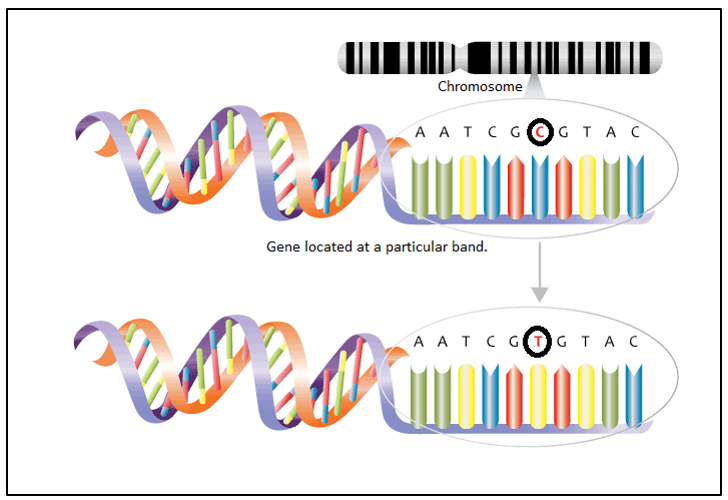
Fig 8: Single nucleotide polymorphism (SNPs) |
Taking the case of sickle cell anemia, where the mutation turned out to be disastrous, sometimes and the mutation can be favorable to the person. The gene present at a particular locus that is made of DNA can bear a single nucleotide polymorphism (SNPs). SNP is a single base-pair change that contributes to the uniqueness of an individual. It varies from individual to individual and can be traced quite easily with recent technologies. It is of various types like addition of a new base-pair, deletion, replacement or frame shift mutations. The significance of SNPs is that a single mismatch in nucleotide sequence predisposes an individual to a particular disease or injury like a ligament tear or can even enhance their traits like increasing their hypoxic resistance and metabolism.
|
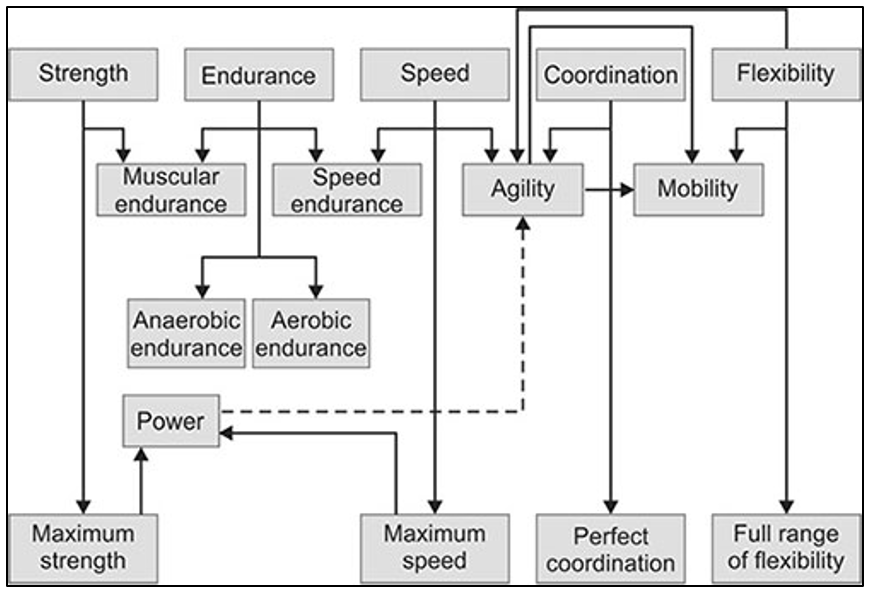
Fig 9: Attributes of Sports Performances.(Geetanjali B, Subhadra M. Nutritional Guidelines for Sportspersons. 10.5005/jp/books/18033_10,2018)https://www.jaypeedigital.com/book/9789352703456) |
The above mentioned concept of SNPs is what makes sportspersons different; in their capacity to perform, excel and heal. Such SNPs often contribute advantageously or negatively to traits like speed, power, endurance, balance, agility, maximal oxygen uptake, anaerobic threshold, anthropometry, flexibility and recovery. That is the reason some sportspersons are skilled at endurance sports, some at power-oriented sports and some can tolerate low oxygen conditions, etc. This is the concept involved in genetic testing of sports performance. Various countries have employed the detection of such SNPs using Genome-Wide Association Studies (GWAS) to characterize the abilities and detect them in upcoming sportspersons as a test of their innate ability to perform.(1)
Based on studies of such single nucleotide polymorphisms in various countries, possibly significant genes that contribute to sports performances, their loci, the attribute and the positive single nucleotide polymorphism and the groups of elite athletes it has been confirmed in has been listed below.
-
- Link between determination of SNPs and Sports Performance
The attributes of sportspersons are encoded by certain select genes. For example: ACTN3 gene that is the alpha-actinin 3 is one of the predominant proteins in α-actinins and is limited to fast-muscle fibers which is responsible for generating force at high velocity. The protein also stabilizes muscle contractile apparatus. At the 577th amino acid position, there is a common replacement of an arginine residue by a stop codon (R to X at 577th position). This allows for only a part of the protein to form due to the premature stop codon (protein synthesis will terminate). Because the complete alpha-actinin was not encoded, the people bearing this mutation tend to tire easily and have slower recovery rates from exercise induced muscle damage.(5) A single replacement of C nucleotide by T nucleotide on the chromosomal locus of 11q13.1, leads to transcription of stop codon thus stopping the protein from being synthesized halfway: an incomplete muscle protein. This negatively affects the sportsperson. In elite Finnish sprint athletes, the replacement was not seen (they bore C nucleotide) and was observed to have the complete protein. This genotype of R577X was seen highly underrepresented in Australian power athletes and none of the Olympian female athletes had XX genotype. (1) (3)
In inference, it could be said that them not having the mutation, enhanced their sports performance.
Similarly, other genetic markers are listed out whose mutations (SNPs) negatively or positively affect the athletic performance.
|
Sl. No. |
Gene Markers |
Locus |
Allele |
Quality |
Countries |
|
1. |
ACTN3 |
11q13.1 |
Rs1815739 C/T |
Power |
Elite Finnish sprint athletes, |
|
2. |
ACE I/D |
17q23.3 |
287bp Alu insertion, rs4646994
|
Endurance, Power |
British and Japanese elite long-distance runners, Australian, Croatian, Russian, and Spanish elite athletes, Italian Olympic endurance athletes, Turkish athletes, Polish rowers. |
|
3. |
PPARA |
22q13.31 |
Power |
Lithuanian power-oriented athletes and they show better hand grip |
|
|
4. |
COL5A1 |
9q32.2-q34.3 |
Rs12722 C/T |
Endurance |
Caucasian Ironman triathletes |
|
5. |
HIF1A |
14q23.2 |
Rs11549465 C/T |
Endurance |
Caucasian male elite athletes, Russian endurance athletes |
|
6. |
NOS3 |
7q36 |
Rs2070744T/C |
Endurance |
Caucasian fastest finishing tri- athletes |
Table 1: List of genetic markers well studied and confirmed in at least three studies. The genetic markers represent the power and endurance phenotype. The required alleles and the functions of the genes are known. (1)
- The biochemistry behind the selection of the following gene markers for sports.
About 11 markers for power and endurance have been confirmed with elite athlete status in more than studies. However, the above-mentioned genes have generated quite an interest and their alleles have been well documented. (1) (27)
Angiotensin Converting Enzyme-I (ACE) gene has multiple functions that boosts the body functions and regulates blood volume and pressure by synthesis of a vasoconstrictor Angiotensin II and degrading vasodilators kinins. The two alleles of interest in the ACE gene are I and D. The I allele boosts endurance performance as it lowers ACE activity thus increasing bradykinin that leads to enhanced substrate delivery to muscles. The D allele, by contrast favors power sports by improving cardiac function. (2)
COL5A1 gene influences the energy cost of running as it encodes a Type V collagen in ligaments.(2) Collagens are a group of extracellular matrix proteins and are most abundant proteins in mammals making up about 25%-35% of the whole-body protein content and the COL5A1 gene encodes the pro-α1 chain of type V collagen. The carriers of rs12722 T polymorphism have more inflexibility that enhances running performance. This was expounded in Caucasian Ironman tri-athletes as the individuals bearing this gene polymorphism ran faster significantly. A second study replicated the results in ultra marathon runners. (9)
Hypoxia-inducible factor-1α (HIF1A) proteins inhibit protein degradation under hypoxic conditions and gets stimulated in turn increasing the rate of erythropoiesis and subsequently increasing oxygen delivery to the muscles. The C allele of HIF1A depicts a higher value of VO2 max which is an important factor governing the capacity of marathon running performance. (2)
HIF1A, NOS3 and PPARA are associated with power status in Ukraine. (27)
ACTN3

Fig 10: Genomic sequence of ACTN3 Gene ID 89 (RefSeqGeneNG_013304.2/ACTN3 transcript variant 1, coding NM_001104.4:c.1729C>T=R[CGA] > Stop [TGA] Source:ncbi.nlm.nih.gov/snp/rs1815739)

Fig 11: Aminoacid sequence encoded by the ACTN3 gene isoform 1 (NCBI Reference Sequence: NP_001095.2) Source: ncbi.nlm.nih.gov/protein/585866354
The predominant genetic variation of ACTN3 is the replacement of arginine by a stop codon. The genotypes in question will be ACTN3 XX, ACTN3 RX and ACTN3 RR. Alpha-actinin 3 is a component of the fast-twitch skeletal muscle fibers and the sequence of ACTN3 that encodes it is strongly conserved. (16) The presence of RR and RX allele encodes the protein completely whereas XX allele means there is a condition of deficiency of alpha-actinin 3 proteins. However, there is no association of XX with any disease as up regulation of its closely related isoform alpha-actinin 2 occurs. (16) The frequency of ACTN3 XX is reduced in athletes from various studies. (12) Though there is no concrete evidence that ACTN3 R allele is associated with the power status, it has been used as a genetic marker by companies.
ACTN3 R577X allele has been linked to exercise-induced muscle damage. (4) In a study, the individuals bearing XX gene had higher pain scores and higher serum CK activity than the RR bearing individuals after performing a series of maximal eccentric knee extensions with both legs concluding that there is a possible protective function of complete α-actinin 3 proteins. (5)Also subjects with RR and RX polymorphisms are more resistant to muscle damage resulting from high intensity genotype than athletes with XX genotype. (14) Due to this endurance ability donated by the R allele, Papadimitrou et al, 2016 demonstrated that male Caucasian sprinters with ACTN3 577RR had the fastest time for 200m and no other candidates with ACTN3 577XX could beat the record. (15)
ACE

Fig 12: NCBI dbSNP query: https://www.ncbi.nlm.nih.gov/snp/rs4646994

Fig 13: NCBI dbSNP query: https://www.ncbi.nlm.nih.gov/snp/rs4646994
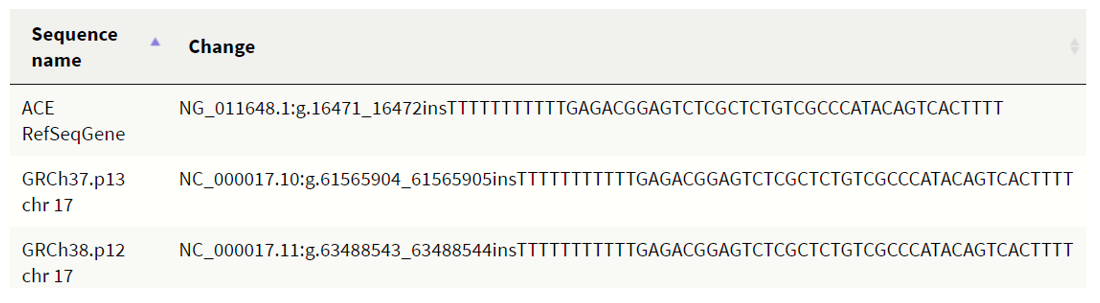
Fig 10 :ACE:IntronVariant; chr17:63488530-63488543 (GRCh38.p12); https://www.ncbi.nlm.nih.gov/snp/rs1799752
The polymorphism is seen in intron 16 of the human ACE gene. The I allele is an insertion of a 287-base pair Alu sequence and is linked to lower circulating tissue ACE while D allele is the deletion of the same. (13)This affects the function of the enzyme activity which is tied to the regulation of blood pressure and cardio-respiratory efficiency. The DD genotype has been associated with power sports and I allele with endurance sports. (8) In fact, ACE DD genotype bearing individuals had fastest 400m sprint time than their ACE II counterparts (15). There is also evidence of ACE II alleles enhancing aerobic power. (26)The presence of the insertion allele has been seen in elite mountaineers, long distance runners, Croatian rowers, Spanish elite athletes, elite Spanish runners, Russian middle-distance runners, Italian Olympic triathlon athletes and also Indian Army tri-athletes. (1)However, exceptions have been noted, for example; Israeli elite marathon runners have DD genotype (24) and hence this association may be dependent on the ethnicity. Also, there are contrary findings and as Papadimitrou et al succinctly put, ACE gene should not be the only factor considered; epigenetic factors like CpG islands, biomechanical factors of the muscles and thropometric factors should be considered. (17)
COL5A
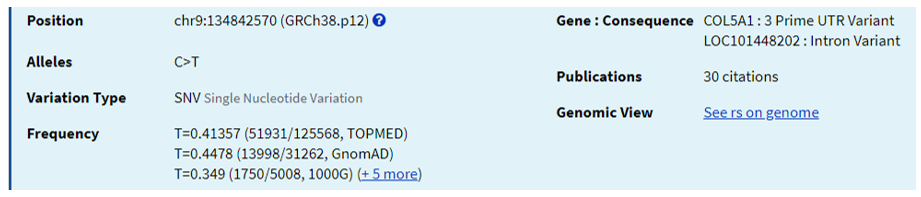

Fig 15: NCBI dbSNP queryhttps://www.ncbi.nlm.nih.gov/snp/rs12722
Apart from inflexibility, COL5A also is used as a marker for muscle injury as it is a genetic component contributing to tendon-ligament injury (TLI). It was observed that COL5A1 polymorphisms lessened the risk of TLI in Caucasians (10). Individuals with CC genotype had a significantly decreased risk of developing chronic Achilles tendinopathy than those with a T allele (28).
HIF1A

Fig 16: NCBI dbSNP queryhttps://www.ncbi.nlm.nih.gov/snp/rs11549465
HIF1A is a transcription factor that regulates many genes in response to hypoxic stimuli and is expressed more in glycolytic muscles. A missense mutation in exon 12 rs11549465 C/T results in substitution of proline with serine at the 582nd position that confers higher stability and transcriptional activity to HIF1A and also promotes glucose metabolism. (7) In subjects with Pro/Pro homozygous genes, it was observed that they can endure longer aerobic exercise whereas the ones with Pro/Ser heterozygous genes and Ser/Ser heterozygous genes endure longer anaerobic workouts. The frequency of HIF1A Ser589 is seen in elite and highly elite weightlifters, Polish, Ukrainian and Russian power-oriented athletes and in one study (11) this allele was linked to an increased proportion of fast-twitch muscle fibers in M.vastus lateralis of all-round speed skaters.
NOS3

Fig 17: NCBI dbSNP Query
Nitric oxide is a signaling molecule involved in activity of human skeletal muscle during exercise and regulating oxygen consumption in skeletal muscles. Genetic variation in Nitric Oxide Synthase gene (NOS3) could be associated with power/sprint performance and was recorded in Ukrainian and Italian power-oriented athletes and Spanish elite power-oriented athletes. The gene polymorphism is ars2070744 T/C substitution that results in Glu298. However, Glu298Asp polymorphism is unfavorable for high sports performance. (1) This is further confirmed by a study of three groups of Spanish Caucasian athletes and in conclusion it was given that the T allele of the -786 T/C polymorphism exercises an advantageous effect on power-oriented sports ability. (6) In case of NOS3, one study (23) proved that physical pre-conditioning evokes a genotype-influenced right ventricular adaptation.
PPARA

Fig 185: NCBI dbSNP Query
Peroxisome proliferator-activated receptor (PPAR) proteins belong to the steroid hormone receptor super family and associate with the retinoid X receptors to form heterodimers that regulate genes involved in lipid and glucose metabolism, adipocyte differentiation, fatty acid transport, carcinogenesis and inflammation. PPARs exist in three different forms –alpha (PPARα), PPAR-beta/delta (PPARβ/δ) and PPAR-gamma (PPARγ), which are encoded by the genes PPARA, PPARD and PPARG. (29-31)
PPARA gene’s role is mainly in encoding proteins that regulate energy homeostasis, lipid metabolism and it is activated under conditions of energy deprivation thus enhancing uptake, utilization and catabolism of fatty acids. It has two genotypes: G and C. Higher proportion of athletes tend to have GG genotype and G allele. (19) Though the PPARA G/C polymorphism is located in the non-coding region of the gene in intron 7 and likely to be non-functional but there is a possibility that this polymorphism is in linkage disequilibrium with a functional variant in the promoter of an enhancer element of the PPARA gene that results in PPARA gene expression. Another possibility is that the PPARA polymorphism interacts with the PPAR receptors and other genes like ApoA1 and ApoB (20-22).
Apart from the above individual genes, certain studies try to elucidate multi-gene models to propose interactions between the various genes. In a study (18), there were interaction effects seen between ACE and NOS3. A three gene model of BDKRB2, AMPD1 and UCP2 showed high amount of accuracy with the results. While the genetic markers would be considered independently, the interactive mechanisms of the action of the genes should not be underestimated.
The most predominantly studied combination of genes is ACE and ACTN3. In competitive cross-country skiing, the allelic advantage was held by individuals bearing ACTN3 RR and ACE ID. (25) A study suggested that the ACTN3 R577X and ACE I/D polymorphisms independently affect the proportion of human skeletal muscle fibers MHC-I and MHC-IIx in men but not in women.(29)
2.7 Genetic testing for sports around the world: Methods and DTCs
The stepwise method of genetic assessment of sportspersons starts by first, the isolation of the genomic DNA content from the swab cells, purification and genetic sequencing. There are established protocols and kits available for this process. DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.(34) The SNP assessment can also be done using Entrez database in the NCBI website called dbSNPs or using a library of artificial DNA.(33)
In vitro diagnostics (IVDs) that are marketed directly to consumers without the involvement of a health care provider are called direct-to-consumer tests (also referred to as DTCs). These tests generally request the consumer collect a specimen, such as saliva or urine, and send it to the company for testing and analysis. Direct-to-consumer testing is expanding the number of people who are able to get genetic testing of their DNA (or genome). This has been increasing in popularity in metropolitan cities for parents who subject their children to such testing to understand if they possess sports talent or not.(35) Such tests are preferred by people due to their non-invasiveness, affectivity and fast results.
SNP assessment fields have grown due to computer chip manufacturers and pharmaceuticals that have partnered together to develop techniques to identify these easily. Microarrays and silicon chips to assess the SNPs are now widely used.
Using the above methods, the analysis of such information is done by Genome Wide Association Studies (GWAS). In this method, scanning of several 100,000 (up to 5 million) markers across complete DNA set is done to check for the performance enhancing polymorphisms (PEPs).(1)
There are two methods of doing these,
- Case-Control Studies: This is the most common study design in sports genomics and here one allele is determined in the DNA sequence and this is checked to be common in a group of elite athletes. To avoid false-positive results, case control studies should have one replication with additional athletic and non-athletic cohorts from different populations.
- Cross-Sectional Association Studies: Here the genotype of an athlete is taken and different traits are measured like VOmax, strength, percentage of fast twitch muscle fibers, cardiac size and lactate formation.
This helps to characterize the indigenous traits of athletes, especially here in India and also provides more scope for a more detailed understanding of the molecular mechanisms of such SNPs.
- DISCUSSION
Athletic performance assessment has always been a crucial step for selection and training. The sports field has come a long way from physical, physiological and biological tests to assess the talent of an upcoming sportsperson. Sports genomics is the new next-level step that has to be taken to get a complete assessment of the individual. The methods to perform this test are not rocket-science, all it requires is a saliva sample and basic biochemical DNA extraction procedures and sequence analysis. The technologies for all this are now widely available and should be made most of. Screening of athletes for their talents can be done in large numbers and at a young age also, as the procedures are user-friendly and the core genetic blueprint never gets modified at any point by natural causes.
While the obtaining of samples has been found to be practical, non-invasive and convenient; the assessment by basic biochemical techniques and next generation sequencing has been established, the genes and SNPs have been characterized by GWAS and there is no dearth of elite athletes and upcoming aspiring athletes- what seems to be a problem?
The main issue that arises here are the ethical concerns. Seema Patel et al (36), proposed guidelines to genetic testing as such screening methods shouldn’t be misused to favor some sportspeople over the others. The screening method should be used to just improve the athlete’s training regimen and check which child has a higher affinity to the specific type of sport (endurance or strength). Jobs and selection of athletes also should not be solely based on this method as athletic status has just 66% of credibility and not 100%. Without proper characterization and biological confirmation of the genetic influence, decisions should not be made. The information obtained should also be in strict confidence of the coach or captain and the athlete in question. The rules and regulations of using this sensitive information has to be firmly established before this method becomes standard procedure.
Other issues that is faced is the lack of characterization of sports performance related SNPs in Indian population. All the above studies have been made in countries other than India and such analysis should also be carried out in India. The ethnicity factor is important and more studies should be conducted on the indigenous people to establish a firm foundation for genetic analysis. Genetic analysts and research scientists should lean into this field to elucidate the molecular mechanisms of favorable SNPs and provide more insights.
Furthermore, identifying a specific gene variant and performing accurate analysis of a specific gene variant allows for incorporation of appropriate lifestyle changes, personalized nutrition and efficient physiotherapy which can enhance athletic performance and effective strategies can be prepared to get the best out of each athlete. Incidents like the deaths due to sickle cell anemia and other genetic disorders can be avoided and athletes can be given individual personalized attention.
Hence in the long term, using this analysis can be a beacon of hope to the sports background of India. Sports has always been celebrated in India since ages, in fact according to the Budget review of India for the financial year 2020-2021, the sports budget has been increased to Rs 2826.92 crores for infrastructure and facilities (37). Pairing such a great boost of encouragement to sports with a concrete sports genetic base can predictably do wonders for the sports performance of Indian sportspersons.
REFERENCES
- Ahmetov II, Fedotovskaya ON. Current Progress in Sports Genomics. 2015. Advanced Clinical Chemistry. 70:247-314
- Moir JM, Kemp R, Folkerts D, Spendiff O, Pavlidis C. Genes and Elite Marathon Running Performance: A Systematic Review. 2019. Journal of Sports Science and Medicine. 18: 559-568.
- Yang N, MacArthur DG, Gulbin JP, Hahn AG, et al. ACTN3 Genotype is associated with Human Elite Performance. 2003. Am J Hum Genet. 73(3): 627-631.
- Pimenta EM, Coelho DB, Veneroso CE, et al. Effect of ACTN3 gene on strength and endurance in soccer players. 2013. J Strength Cond Res. 27(12): 3286-3292.
- Vincent B, Windelinckx A, Nielens H, Ramaekers M, et al. Protective role of alpha-actinin-3 to an acute eccentric exercise bout. 2010. J Appl Physiol. 109(2): 564-573.
- Gomez-Gallego F, Ruiz RJ, Buxens A, Santiago C, et al. The -786 T/C Polymorphism of Nos3 Gene Is Associated with Elite Performance in Power Sports. 2009. Eur J Appl Physiol. 107(5): 565-569.
- Gabbasov RT, Arkhipova AA, Borisov AV et al. The HIF1A gene Pro582Ser polymorphism in Russian strength athletes. 2013. J Strength Cond Res. 27(8): 2055-2058.
- Yang Y, Ma F, Li X, Zhou F, Gao C, et al. The Association of Sport Performance with ACE and ACTN3 Genetic Polymorphisms: A Systematic Review and Meta-Analysis. 2013. PLoS One. 8(1): e54685.
- Posthumus M, Schwellnus MP, Collins M. The COL5A1 gene: a novel marker of endurance running performance. 2011. Med Sci Sports Exerc. 43(4):584-589.
- Pabalan N, Tharabenjasin P, Phababpha S, Jarjanazi H. Association of COL5A1 gene polymorphisms and risk of tendon-ligament injuries among Caucasians: a meta analysis. 2018. Sports Med (Open). 4:46.
- Ahmetov II, HakimullinaAM, Lyubaeva EV, Vinogradova OL, Rogozkin VA. Effect of HIF1A gene polymorphism on human muscle performance. 2008. Bull ExpBiol Med. 146(3): 351-353.
- Roth SM, Walsh S, Liu D, Metter EJ, Ferrucci L, Hurley BF. The ACTN3 R577X nonsense allele is underrepresented in elite-level strength athletes. 2008. Eur J Hum Genet. 16(3):391-4.
- Chiu YH, Lai JI, Tseng CY, Wang SH, Li LH, Kao WF, How CK, et al. Impact of angiotensin I converting enzyme gene I/D polymorphism on running performance, lipid, and biochemical parameters in ultra-marathoners. 2019. Medicine (Baltimore). 98(29): e16476.
- Pimenta EM, Coelho DB, Cruz IR, et al. The ACTN3 genotype in soccer players in response to acute eccentric training. 2012. Eur J Appl Physiol. 112: 1495-1503.
- Papadimitrou DI, Lucia A, Pitsiladis PY, et al. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: a multicohort study. 2016. BMC Genomics. 17: 285.
- Kikuchi N and Nakazoto K. Effective utilization of genetic information for athletes and coaches: focus on ACTN3 R577X polymorphism. 2015. J ExercNutrBiochem. 19(3): 157-164.
- Papadimitrou DI, Lockey JS, Voisin S, Herbert AJ et al. No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. 2018. BMC Genomics. 19:13.
- Gronek P, Gronek J, Spieszny M, Niewcas M, et al. Polygenic Study of Endurance Associated Genetic Markers NOS3 (Glu298Asp), BDKRB2 (-9/+9), UCP2 (Ala55Val), AMPD1 (Gln45Ter) and ACE (I/D) in Polish Male Half Marathoners. 2018. J Hum Kinetics. 64:87-98.
- Lopez-Leon S, Tuvblad C and Forero DA. Sports genetics: the PPARA gene and athletes’ high ability in endurance sports. A systematic review and meta-analysis. 2016. Sport. 33:3-6.
- Liu M, Zhang J, Guo Z, Wu M, Chen Q, Zhou Z, Ding Y, Luo W. Association and interaction between 10 SNP of peroxisome proliferator-activated receptor and non-HDL-C. 2015. Zhonghua Yu Fang Yi XueZaZhi. 49(3):259-64.
- Hai B, Xie HJ, Guo ZR, Wu M, Chen Q, Zhou ZY, Yu H, Ding Y. [Association of both peroxisome proliferator-activated receptor, gene-gene interactions and the lipid accumulation product]. 2013. Zhonghua Liu Xing Bing XueZaZhi. 34(11):1071-6.
- Luo W, Guo Z, Wu M, Hao C, Hu X, Zhou Z, Zhou Z, Yao X, Zhang L, Liu J. Association of peroxisome proliferator activated receptor alpha/delta/gamma with obesity, and gene-gene interaction, in the Chinese Han population. 2013. J Epidemiol. 23(3):187-94.
- Szelid Z, Lux A, Kolossvary M, Toth A, et al. Right ventricular adaptation is associated with Glu298Asp variant of the NOS3 gene in elite athletes. 2015.PLoS ONE. 10(10): e0141680.
- Amir O, Amir R, Yamin C, Attias E, et al. The ACE deletion allele is associated with Israeli elite endurance athletes. 2007. Exp Physiol. 92.5: 881–886.
- Magi A, Unt E, Prans E, Raus L, et al. The Association Analysis between ACE and ACTN3 Genes Polymorphisms and Endurance Capacity in Young Cross-Country Skiers: Longitudinal Study. 2016. Journal of Sports Science and Medicine. 15:287-294.
- Bueno S, Pasqua LA, de Arau ´jo G, Eduardo Lima-Silva A, Bertuzzi R. The Association of ACE Genotypes on Cardiorespiratory Variables Related to Physical Fitness in Healthy Men. 2016. PLoS ONE. 11(11): e0165310.
- Drozdovska SB, Dosenko VE, Ahmetov II, Ilyin VN. The association of gene polymorphisms with athlete status in Ukrainians. 2013. Biol Sport. 30:163-167.
- Maffulli N, Marglott K, Longo UG. The genetics of sports injuries and athletic performance. 2013. Muscles, Ligaments and Tendons Journal. 3(3): 173-189.
- Kumagai H, Tobina T, Kakigi R, Tsuzuki T, Zempo H, et al. Role of selected polymorphisms in determining muscle fiber composition in Japanese men and women. 2018. J Appl Physiol. 124(5): 1377–1384.
- Dubuquoy L,Dharancy S,Nutten S,Pettersson S,Auwerx J,Desreumaux P. Role of peroxisome proliferator-activated receptor γ and retinoid X receptor heterodimer in hepato-gastroenterological diseases. 2002. Lancet. 360:1410–1418.
- CabreroA, Laguna J, VazquezM. Peroxisome proliferator-activated receptors and the control of inflammation. 2002. Curr. Drug Targets-Inflamm Allergy, 1:243–248.
- Petr M, Petr S, et al. The Role of Peroxisome Proliferator-Activated Receptors and Their Transcriptional Coactivators Gene Variations in Human Trainability: A Systematic Review. 2018. Int. J. Mol. Sci. 19:1472.
- McArdle WD, Katch FI, Katch VL. Exercise Physiology. Lippincott, Williamsand Wilkins, 7th Ed:940-944, 2008.
- Behjati S, Tarpey PS. What is next generation sequencing? Archives of Disease in Childhood. Education and Practice. 98 (6): 236–37.
- Genetic Sports Gene ACTN3 DNA Test DNA Test (https://dnalabsindia.com/dna-genetic-sports-gene-actn3-dna-test-cms-19.html)
- Patel, S., & Varley, I. (2019). Exploring the Regulation of Genetic Testing in Sport. The Entertainment and Sports Law Journal, 17(1), 5.
- https://economictimes.indiatimes.com/news/sports/government-allocates-rs-2826-92-crore-to-sports-budget-rs-50-crore-increase-from-last-year/articleshow/73839805.cms

















